Zymolyase®
Efficient degradation of yeast cell walls
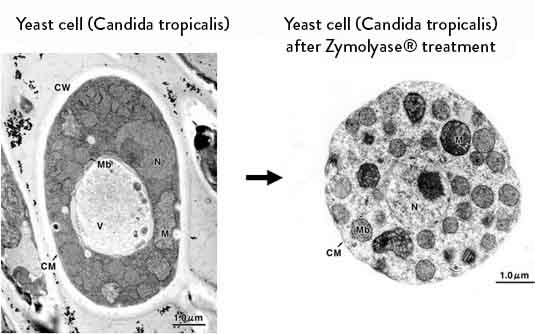
The digestion of fungal and yeast cell walls is a necessary part of the protocol for many experimental procedures including immunofluorescence, transformation, protein purification and spheroplasting. To achieve this digestion, lytic enzymes such as Zymolyase are routinely used.
AMSBIO offers Zymolyase, produced by a submerged culture of Arthrobacter luteus1, which has an effective lytic activity against cell walls of viable yeast cells2,3 to produce protoplast or spheroplast of various strains of yeast cells. An essential enzyme for the lytic activity of Zymolyase is β-1,3-glucan laminaripentaohydrolase. It hydrolyzes linear glucose polymers at β -1,3-linkages and releases specifically laminaripentaose as the main and minimum product unit4,5,10,11.
Benefits
- Convenience: Provided as a lyophilized powder and reconstituted with a buffer solution
- Very efficient cell wall digestion
- Prepared from Arthrobacter luteus, their essential enzyme activity is β-1,3-glucan laminaripentao-hydrolase
Applications
- Protoplast/spheroplast preparation
- Yeast cell fusion
- Transformation of yeast cells
- Yeast genetics
There are two preparations of Zymolyase: Zymolyase-20T and Zymolyase-100T, having lytic activity of 20,000 units/g and 100,000 units/g respectively. Zymolyase-20T is ammonium sulfate precipitate while Zymolyase-100T is a further purified preparation by affinity chromatography9. Lytic activity varies depending on yeast strain, growth stage of yeast, or cultural conditions. Further information related to Zymolyase® can be obtained in the reference section below12-16.
Supporting Documents
Specifications
| Form | Lyophilized powder | ||
| Purification | Zymolyase® -20T:(NH4)2SO4 precipitation Zymolyase® -100T:Affinity Chromatography | ||
| Activity | Zymolyase® -20T:20,000 units/gram Zymolyase® -100T:100,000 units/gram | ||
| Essential enzyme | β.-1, 3-glucan laminaripentaohydrolase | ||
| Other activities contained | Zymolyase® - 20T | Zymolyase® - 100T | |
| B-1, 3-glucanase: | ca. 1.5 x 106 units/g | ca. 1.0 x 107 units/g | |
| protease: | ca. 1.0 x 104 units/g | ca. 1.7 x 104 units/g | |
| mannanase: | ca. 1.0 x 106 units/g | ca. 6.0 x 104 units/g | |
| Contaminants: | Trace amounts of amylase, xylanase, phosphatase No DNase, RNase detected | ||
| Stable pH | 5~10 | ||
| Heat stability | The lytic activity is lost on incubation at 60°C for 5 minutes | ||
| Specificity (lytic spectrum): | Ashbya, Candida, Debaryomyces, Eremothecium, Endomyces, Hansenula, Hanseniaspora, Kloeckera, Kluyveromyces, Lipomyces, Metschikowia, Pichia, Pullularia, Torulopsis, Saccharomyces, Saccharomycopsis, Saccharomycodes, Schwanniomyces, etc. | ||
| Activators: | SH compound such as cystein, 2-mercaptoethanol of dithiothreitol | ||
| Stability: | No loss of activity was found after storage for 1 year at 4°C | ||
Citations
Domestication and divergence of Saccharomyces cerevisiae beer yeasts.
Gallone, B., et al (2016) Cell, 166(6), 1397-1410.
Keeping Candida commensal: how lactobacilli antagonize pathogenicity of Candida albicans in an in vitro gut model
Graf, K., Last, A., Gratz, R., Allert, S., Linde, S., Westermann, M., ... & Hube, B. (2019). Disease models & mechanisms, 12(9), dmm039719.
Structure-based directed evolution improves S. cerevisiae growth on xylose by influencing in vivo enzyme performance.
Lee, M., Rozeboom, H. J., Keuning, E., de Waal, P., & Janssen, D. B. (2020). Biotechnology for Biofuels, 13(1), 5.
Crosstalk between chromatin structure, cohesin activity and transcription.
Maya-Miles, D., Andújar, E., Pérez-Alegre, M., Murillo-Pineda, M., Barrientos-Moreno, M., Cabello-Lobato, M. J., ... & Prado, F. (2019). Epigenetics & chromatin, 12(1), 47.
Molecular Wiring of a Mitochondrial Translational Feedback Loop.
Salvatori, R., Kehrein, K., Singh, A. P., Aftab, W., Möller-Hergt, B. V., Forne, I., ... & Ott, M. (2019). Molecular Cell.
The Aspartic Protease Ddi1 Contributes to DNA-Protein Crosslink Repair in Yeast.
Serbyn, N., Noireterre, A., Bagdiul, I., Plank, M., Michel, A. H., Loewith, R., ... & Stutz, F. (2020). Molecular Cell.
Tor-mediated induction of autophagy via an Apg1 protein kinase complex.
Kamada, Y., et al (2000). The Journal of Cell Biology, 147(2), 435-446.
Formation process of autophagosome is traced with Apg8/Aut7p in yeast.
Kirisako, T., et al (1999). The Journal of Cell Biology, 147(2), 435-446.
Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein.
Lopez-Girona, A., et al (1999). Nature, 397(6715), 172-175.
Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins.
Krobitsch, S., & Lindquist, S. (2000). PNAS, 97(4), 1589-1594.
Construction and characterization of a yeast artificial chromosome library containing seven haploid human genome equivalents.
Albertsen, H.M., et al (1990). PNAS, 87(11), 4256-4260.
Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis.
Lippincott, J., & Li, R. (1998). The Journal of Cell Biology, 140(2), 355-366.
ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1.
You, Z., et al (2005). Molecular and Cellular Biology, 25(13), 5363-5379.
SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases.
Guo, S., et al (1999). JBC, 274(19), 12990-12995.