Exosome Nucleic Acid Isolation
Extracellular vesicles (EV) transport functional RNA molecules in the target cell. EV-derived miRNAs (typically pathogenic miRNAs) can be exploited as novel therapeutic targets or disease biomarkers.
miRNAs seem to play critical roles as transcriptional and post-transcriptional regulators of epigenetic mechanisms and cell processes.
They have been linked to the etiology, progression and prognosis of cancer. Similar miRNA expression patterns between tumour tissue samples and circulating exosomes have been observed.
ExoRNA
Our kit allows for RNA extraction from re-isolated exosomes (ultracentrifugation, chemical precipitation, immunocapture, size exclusion chromatography).

Applications
- Direct capture of EVs and exosome RNA extraction from human biofluids and cell culture media without the need for any initial exosome purification step.
- Simultaneous miRNA and mRNA profiling (qRT-PCR, RT-PCR and microarray).
Advantages
- High yield of total RNA (including small RNAs)
- Fast, user-friendly protocol
- Small starting amount of sample required (less than 1 ml)
ExoRNA Products
| Name | Datasheet | Packsize | Order |
|---|---|---|---|
| RNA Basic Kit | 25 reactions | View |
ExoRNA vs. Competitor
The efficiency of our ExoRNA kit was tested vs a competitor kit for RNA extraction from plasma derived EVs.
Our Method
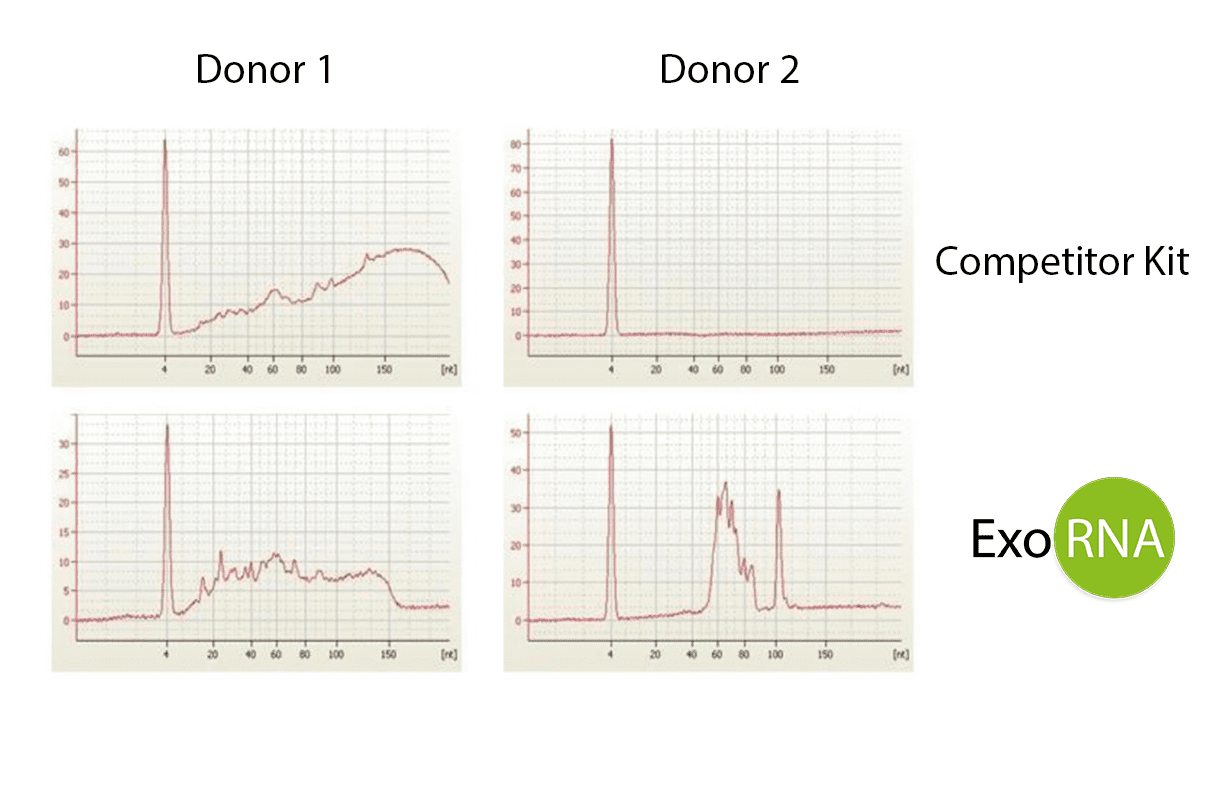
Extraction of total exosome RNA was performed using 100 µl of healthy donor plasma from two different donors (Donor 1 and Donor 2). The competitor’s kit and our ExoRNA kit were both used on each donors’ plasma. RNA quality was evaluated by electropherogram (Figure 1) with Small RNA microfluidic chips (Agilent 2100 Bioanalyzer).
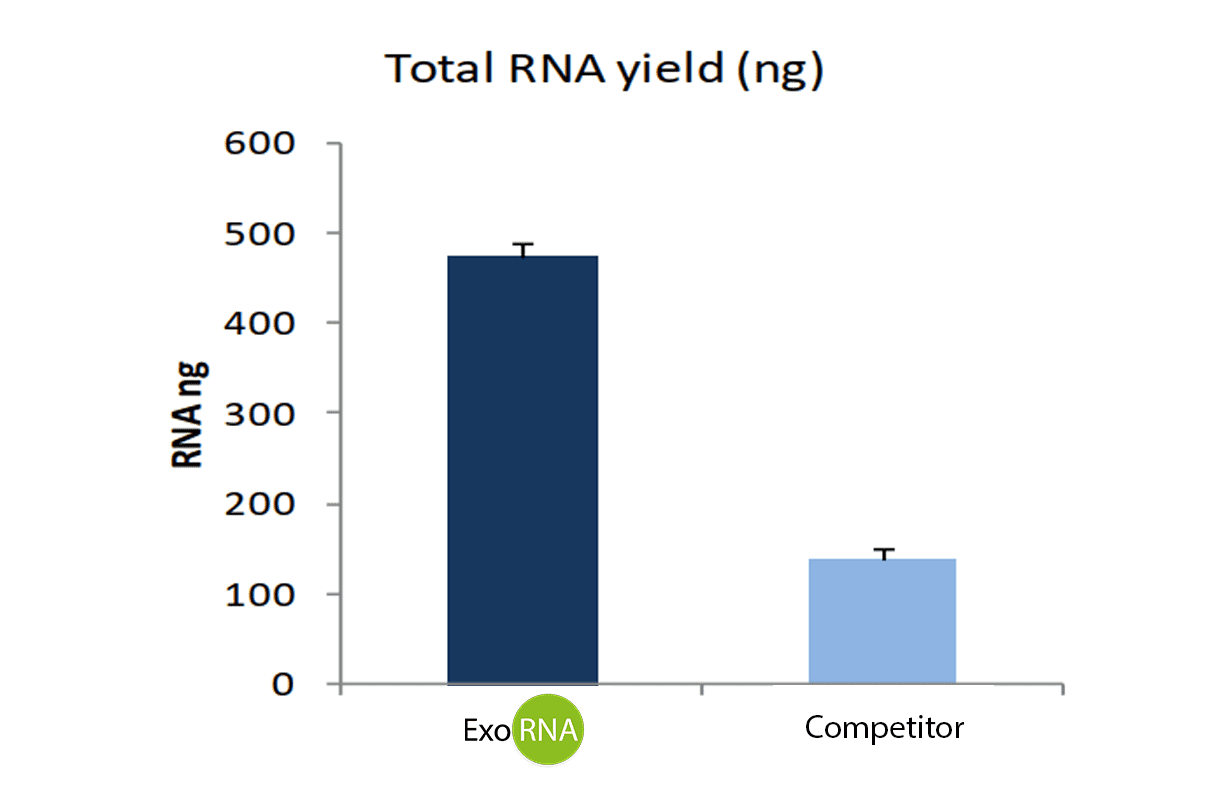
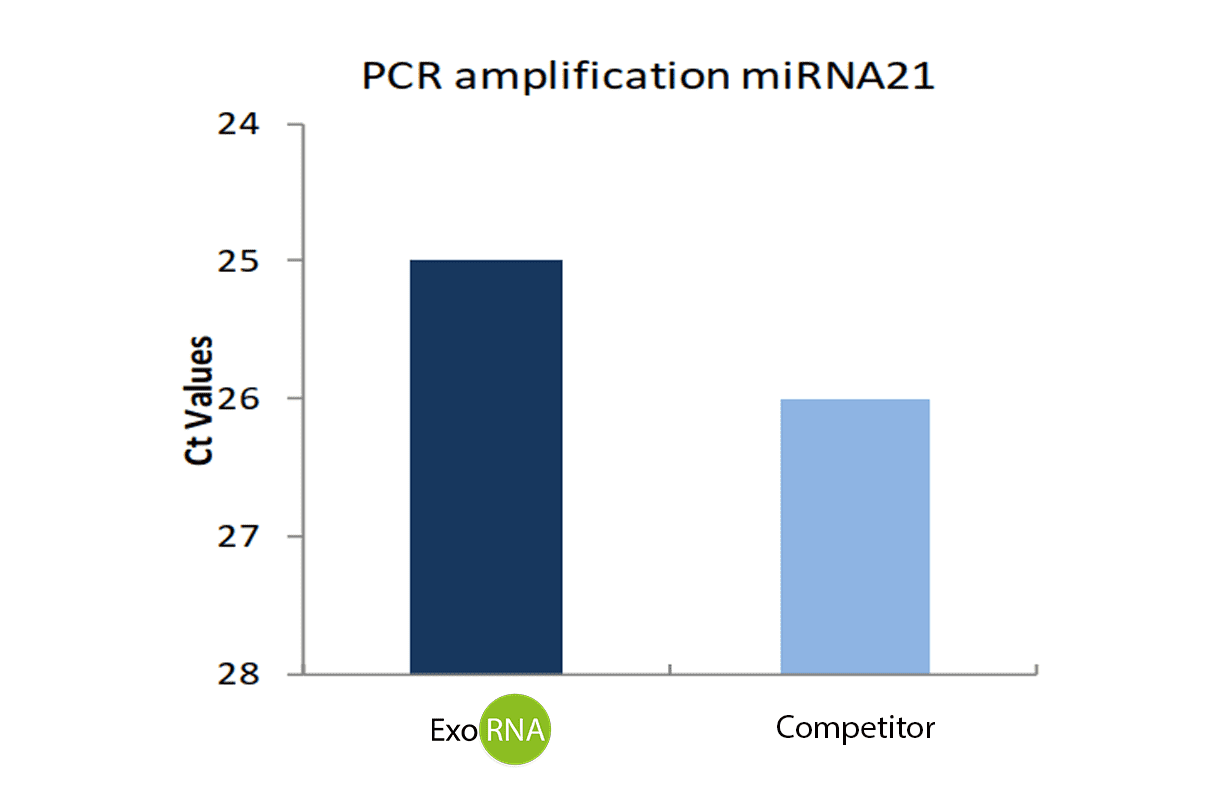
RNA yield was quantified by Nanodrop (Figure 2) and extracted RNA was then retrotranscribed using the miScript II RT kit (Qiagen). miR-21 marker was amplified by qPCR (Fig 3).